Please select your location and preferred language where available.
Challenging to the Brain with Data Storage
KIOXIA Corporation conducts joint collaboration research with the Synthetic
Neurobiology group at the MIT Media Lab on signal processing systems for neuroscience utilizing high-speed, high-capacity SSDs
KIOXIA Corporation (KIC) has been building signal processing systems for neuroscience, taking advantage of high-speed, high-capacity solid-state drives (SSD), in collaboration with the Synthetic Neurobiology group at the MIT Media Lab [1]. To accelerate this collaboration, our company has been sending a visiting scientist to this group since December of 2015.
The Synthetic Neurobiology group, led by Prof. Edward Boyden, is developing technologies for high-precision control and observation of the brain, with the goal of understanding how the brain works at the level of its fundamental constituents such as neurons, and applying the acquired knowledge towards treatment for neurological disorders. KIC is working on accelerating the analyses of the large volume of data generated by these technologies (Fig. 1).
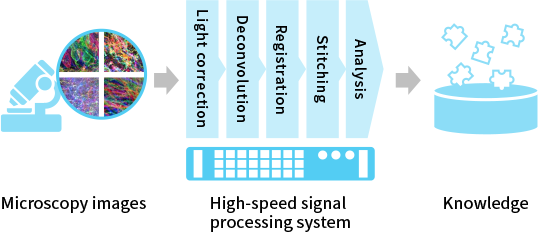

To analyze a huge amount of data
For instance, the Synthetic Neurobiology group has invented a method called Expansion Microscopy [2], where a specimen is embedded in a swellable polymer mesh and is physically expanded, enabling detailed imaging of the brain at a resolution as high as tens of nanometers using conventional light microscopes. The current data size is already a few terabytes per experiment, and it is expected to keep increasing.
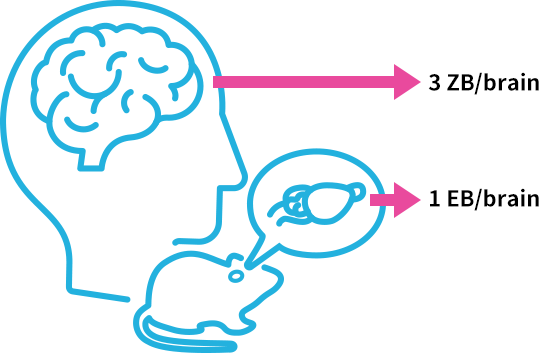
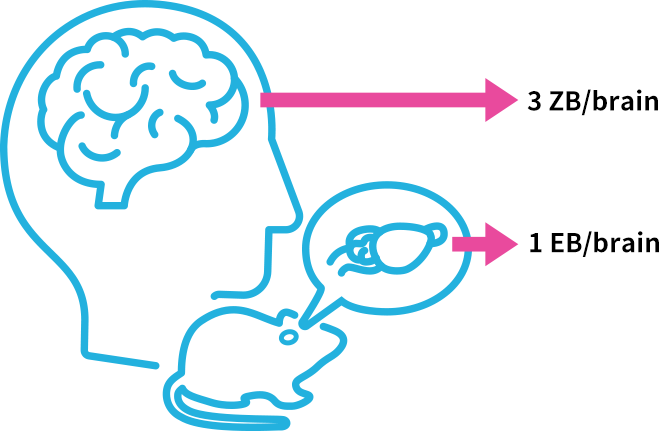
A whole mouse brain can be translated into one exabyte, and a whole human brain can be converted to three zettabytes [3]. The required processing speed is one gigabyte per second currently and will be over one terabyte per second in the future. Efficient processing and visualization of large data is one of the major challenges in conducting research. With the goal of accelerating research cycles through data analysis, KIC contributes to cutting-edge research by building high-speed signal processing systems effectively combining parallel processing devices such as CPUs and GPUs [4] with SSDs, devices which accelerate data reading and writing.
Realizing the real-time visualization of data from a mouse hippocampus
As an example of real-time visualization of microscopy data, the Synthetic Neurobiology group has captured microscopy images of a slice of the mouse brain’s hippocampus using Expansion Microscopy [5], and our company digitally reconstructed a three-dimensional (3D) volume from the images and visualized it on a computer. The dataset is as large as 5 terabytes. The specimen initially measuring 1.2 mm long x 0.6 mm wide x 0.2 mm thick was expanded 4.5 times (around 100 times in volume), and was then imaged multiple times with a light sheet microscope to produce about 340,000 high-resolution images where one pixel corresponds to 50 nanometers (Fig. 2).
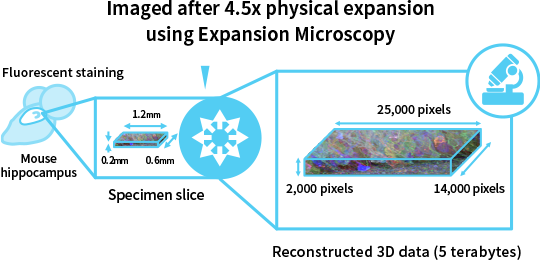

The reconstructed dataset consists of 700 billion (25,000 x 14,000 x 2,000) data points. In order to visualize such large-scale 3D data interactively from a viewpoint that the researcher desires, high-capacity, high-speed SSDs are indispensable. Our company developed a rendering algorithm involving transferring data required for the specified viewpoint rapidly from SSDs to GPUs, achieving real-time rendering performance. In collaboration with NHK Japan Broadcasting Corporation, our company is also working on visualizing the data on an 85-inch large screen at 8K (7,680 x 4,320 pixels) Super Hi-Vision resolution (Fig. 3) [6].

Using state-of-the-art microscopes to visualize the entire brain of a fruit fly in video
Moreover, with the use of a state-of-the-art lattice light sheet microscope, an expanded whole brain of a fruit fly was imaged at an effective pixel size of 24 nanometers [7], and KIC has contributed to making a video visualizing these 11 terabytes of data.
by Y. Bando (KIOXIA Corp/MIT), K. Hiwada (KIOXIA Corp), R. Gao (MIT/Janelia), S. Upadhyayula (HMS/Janelia), S. Asano (MIT), Y. Aso (Janelia), E. Betzig (UC Berkeley/Janelia), E. Boyden (MIT), unmodified from the original,
licensed under CC BY 4.0.
In addition to massively parallel data processing using CPUs, GPUs and SSDs as seen in the visualization example above, KIOXIA Corporation aims at further acceleration of large-volume data processing through the development of novel hardware accelerators and ultrafast interfaces, and thereby contributing to the information society.
[1] Synthetic Neurobiology Group website
[2] Chen, Tillberg, Boyden. Expansion Microscopy, Science 347(6221):543-548, 2015
[3] The values are calculated under the assumption that the average brain volumes (mouse 500 mm3, human 1400 cm3) are digitized at 20 nanometers in 8 colors with 16 bits per color. One exabyte is 1018 bytes, and one zettabyte is 1021 bytes.
[4] CPU: Central Processing Unit, GPU: Graphics Processing Unit (used not only for graphics but also for acceleration of general arithmetic operations)
[5] Asano et al. Expansion Microscopy: Protocols for Imaging Proteins and RNA in Cells and Tissues, Current Protocols in Cell Biology 80(1):e56, 2018
[6] Bando et al. Interactive 3D Visualization of Terabyte-sized Nanoscale Brain Images at 8K Resolution, Neuroscience 2017 Abstract 531.11, 2017
[7] Gao et al. Cortical Column and Whole-Brain Imaging with Molecular Contrast and Nanoscale Resolution, Science 363(6424):eaau8302, 2019
This article was published in June 2019.
The information on this page is true and accurate at the time of publication.
